Benutzer:LinoSiR/Oxidosqualen-Zyklase
Wenn du dies liest:
|
Wenn du diesen Artikel überarbeitest:
|
Oxidosqualene cyclases (OSC) are enzymes involved in cyclization reactions of 2,3-oxidosqualene to form sterols or triterpenes.[1]
Introduction
There are two major groups of sterol-producing OSC enzymes:
- Cycloartenol synthase (CAS), found in all plants, which produces primarily cycloartenol
- Lanosterol synthase (LAS), found in all animals and fungi, and occasionally in plants, which produces primarily lanosterol
Sterols and triterpenes are extremely diverse classes of natural products, particularly in plants, which often contain numerous OSC enzymes with different substrate and product specificities;[1] common examples include lupeol synthase and beta-amyrin synthase.[2] OSC enzymes' catalytic mechanism is similar to the prokaryotic squalene-hopene cyclase.[3]
Directed evolution and protein design have been used to identify small numbers of point mutations that alter the product specificities of OSC enzymes, most notably in altering a cycloartenol synthase to produce predominantly lanosterol.[4]
Structure
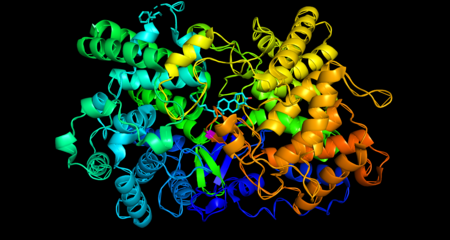
Oxidosqualene cyclase is a monomeric enzyme. Its active site consists of a depression between two barrel domains.[5] The active site is mostly made up of acidic amino acids in the majority of organisms.[6] The residues in the active site make it energetically favorable for oxidosqualene to take on a more folded conformation, which closely resembles its product.[5] This crucially sets the substrate up for the series of reactions that form the rings. Oxidosqualene is located in the cell’s microsome membranes where it can easily harvest its hydrophobic substrate and turn out its hydrophobic product.[7]
Biological Function
Oxidosqualene cyclase is a key enzyme in the cholesterol biosynthesis pathway. It catalyzes the formation of lanosterol, which is then converted through many steps into cholesterol. The body uses cholesterol for temperature regulation. It is also a precursor for testosterone in males and oestradiol in females.[8]
Regulation
The enzyme’s genetic expression is regulated by sterol regulatory element binding protein (SREBP-2), a molecule which also regulates the expression of other enzymes in the cholesterol biosynthesis pathway.[9]
Enzyme Mechanism
Mechanistically, the enzyme catalyzes the formation of four rings along the long chain of the substrate (oxidosqualene), producing lanosterol. This cyclization is one of the most complex known enzyme functions[10] and is highly selective.[11] In the enzyme’s active site, a histidine residue activates an aspartic acid residue, which protonates the substrate’s epoxide, setting off a series of carbon-carbon bond formations that form rings.[7][12] Finally, the enzyme deprotonates to yield lanosterol, which has a hydroxyl group instead of an epoxide. This hydroxyl group can be seen in the image above.
Disease Relevance
High blood cholesterol, also called hypercholesterolemia, significantly increases the risk of stroke, heart attack, and peripheral artery disease. If untreated, it can also lead to plaque accumulation in blood vessels, which is known as atherosclerosis.[13] For this reason, the sterol biosynthetic pathway has long been a target for the drug development industry. Statins, which inhibit HMG-CoA reductase (an enzyme that catalyzes an earlier step in the cholesterol biosynthesis pathway) are commonly prescribed to treat high cholesterol. However, the efficacy and safety of statins has been recently scrutinized in a number of reports.[14][15][16] This is largely because blocking the cholesterol biosynthesis pathway before squalene has been found to disrupt the synthesis of isoprenoids, which are used for the biosynthesis of key molecules in RNA transcription, ATP synthesis, and other essential cell activities.[17] Oxidosqualene cyclase, which is downstream of squalene in the pathway, is an attractive target for inhibition. Many inhibitors have been proposed, among them steroid analogs, phenol-based compounds, benzamide and carboxamide derivatives, and nitrogen-containing heterocyclic compounds. The most effective inhibitors have a hydrogen-bond acceptor at a specific distance away from a hydrophobic region.[6] Inhibitors of oxidosqualene cyclase have shown promise as antimicrobial agents as well, because they’ve been shown to kill off trypanosoma cruzi.[18][19] Trypanosoma cruzi is a parasite transmitted to people by insects, mostly in Latin America. The parasite causes a disease called Chagas disease, in which acute infections around an insect bite can lead to more serious complications, such as decreased heart, esophagus, colon, and even brain function.[20]
Evolution
Stork, et al. compared the protein sequences of C. albicans oxidosqualene cyclase with the analogous enzyme (squalene cyclase) in two different bacteria and found conserved regions in the former.[21] Rabelo et al. found a conserved active site across seven organisms.[6] It is believed that animal and fungal oxidosqualene cyclases likely evolved from their prokaryotic counterparts.[21]
References
External links
[[Category:Isomerases]]
- ↑ a b Ramesha Thimmappa, Katrin Geisler, Thomas Louveau, Paul O'Maille, Anne Osbourn: Triterpene Biosynthesis in Plants. In: Annual Review of Plant Biology. 65, Nr. 1, 29 April 2014, S. 225–257. doi:10.1146/annurev-arplant-050312-120229. PMID 24498976.
- ↑ S. Sawai: Plant Lanosterol Synthase: Divergence of the Sterol and Triterpene Biosynthetic Pathways in Eukaryotes. In: Plant and Cell Physiology. 47, Nr. 5, 15 March 2006, S. 673–677. doi:10.1093/pcp/pcj032. PMID 16531457.
- ↑ KU Wendt, K Poralla, GE Schulz: Structure and function of a squalene cyclase.. In: Science. 277, Nr. 5333, 19 September 1997, S. 1811–5. doi:10.1126/science.277.5333.1811. PMID 9295270.
- ↑ Silvia Lodeiro, Tanja Schulz-Gasch, Seiichi P. T. Matsuda: Enzyme Redesign: Two Mutations Cooperate to Convert Cycloartenol Synthase into an Accurate Lanosterol Synthase. In: Journal of the American Chemical Society. 127, Nr. 41, October 2005, S. 14132–14133. doi:10.1021/ja053791j. PMID 16218577.
- ↑ a b Lord of the rings - the mechanism for oxidosqualene:lanosterol cyclase becomes crystal clear. In: Cell. 26, Nr. 7, 13 June 2005, S. 335–340. doi:10.1016/j.tips.2005.05.004. PMID 15951028.
- ↑ a b c Design strategies of oxidosqualene cyclase inhibitors: Targeting the sterol biosynthetic pathway. In: The Journal of Steroid Biochemistry and Molecular Biology. 171, July 2017, S. 305–317. doi:10.1016/j.jsbmb.2017.05.002. PMID 28479228.
- ↑ a b PDB101: Molecule of the Month: Oxidosqualene Cyclase. In: RCSB: PDB-101 . Abgerufen am 9. März 2019.
- ↑ Berg JM, Tymoczko JL, Stryer L (2012). Biochemistry (7th ed.). New York: W.H. Freeman and Company. Vorlage:ISBN.
- ↑ Joseph L. Goldstein, Michael S. Brown, Sahng Wook Park, Norma N. Anderson, Janet A. Warrington, Nila A. Shah, Jay D. Horton: Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. In: Proceedings of the National Academy of Sciences. 100, Nr. 21, 14. Oktober 2003, ISSN 0027-8424, S. 12027–12032. bibcode:2003PNAS..10012027H. doi:10.1073/pnas.1534923100. PMID 14512514. PMC 218707 (freier Volltext).
- ↑ Gurr, M. I.; Harwood, J. L. (1991), “Metabolism of structural lipids”, in Lipid Biochemistry: An Introduction, Springer US, DOI:, ISBN 9781461538622, pages 295–337
- ↑ Bloch, Konrad. “The Biological Synthesis of Cholesterol.” Nobel Lecture, December 11, 1964.
- ↑ Armin Ruf, Martine Stihle, Michael Hennig, Henrietta Dehmlow, Johannes Aebi, Jörg Benz, Brigitte D'Arcy, Tanja Schulz-Gasch, Ralf Thoma: Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. In: Nature. 432, Nr. 7013, November 2004, ISSN 1476-4687, S. 118–122. bibcode:2004Natur.432..118T. doi:10.1038/nature02993. PMID 15525992.
- ↑ High Blood Cholesterol | National Heart, Lung, and Blood Institute (NHLBI). In: www.nhlbi.nih.gov . Abgerufen am 9. März 2019.
- ↑ James M. Wright, Nicholas Jewell, Harriet G. Rosenberg, John D. Abramson: Should people at low risk of cardiovascular disease take a statin?. In: BMJ. 347, 22. Oktober 2013, ISSN 1756-1833, S. f6123. doi:10.1136/bmj.f6123. PMID 24149819.
- ↑ David M. Diamond, Uffe Ravnskov: How statistical deception created the appearance that statins are safe and effective in primary and secondary prevention of cardiovascular disease. In: Expert Review of Clinical Pharmacology. 8, Nr. 2, March 2015, ISSN 1751-2441, S. 201–210. doi:10.1586/17512433.2015.1012494. PMID 25672965.
- ↑ Terje R. Pedersen, Jonathan A. Tobert: Benefits and Risks of HMG-CoA Reductase Inhibitors in the Prevention of Coronary Heart Disease. In: Drug Safety. 14, Nr. 1, 1. Januar 1996, ISSN 1179-1942, S. 11–24. doi:10.2165/00002018-199614010-00003. PMID 8713485.
- ↑ Patrick Casey: Biochemistry of protein prenylation. In: Journal of Lipid Research. 33, October 19, 2017, S. 1731–1739.
- ↑ Jerald C. Hinshaw, Dae-Yeon Suh, Philippe Garnier, Frederick S. Buckner, Richard T. Eastman, Seiichi P. T. Matsuda, Bridget M. Joubert, Isabelle Coppens, Keith A. Joiner: Oxidosqualene cyclase inhibitors as antimicrobial agents. In: Journal of Medicinal Chemistry. 46, Nr. 20, 25. September 2003, ISSN 0022-2623, S. 4240–4243. doi:10.1021/jm034126t. PMID 13678402.
- ↑ Wesley C. Van Voorhis, Aaron J. Wilson, John H. Griffin, Frederick S. Buckner: Potent Anti-Trypanosoma cruzi Activities of Oxidosqualene Cyclase Inhibitors. In: Antimicrobial Agents and Chemotherapy. 45, Nr. 4, 1. April 2001, ISSN 0066-4804, S. 1210–1215. doi:10.1128/AAC.45.4.1210-1215.2001. PMID 11257036. PMC 90445 (freier Volltext).
- ↑ CDC-Centers for Disease Control and Prevention: CDC - Chagas Disease - Disease. In: www.cdc.gov . 2. Mai 2017. Abgerufen am 9. März 2019.
- ↑ a b Gilbert Stork, A. W. Burgstahler: The Stereochemistry of Polyene Cyclization. In: Journal of the American Chemical Society. 77, Nr. 19, 1. Oktober 1955, ISSN 0002-7863, S. 5068–5077. doi:10.1021/ja01624a038.